1. Introduction#
This article records two methods of PCA analysis using Python, and visualizes 2-dimensional results
1.1 What’s PCA?#
When it comes to methods of reducing dimension, PCA that is an unsupervised linear transformation technique, must be not ignored. Moreover, if you want to know the subtle relationships among data set and reduce the computational complexity in downstream analysis, the PCA may be your best choice! Meanwhile, if you would like to present your data in a 2-dimension or 3-dimension coordinate system, and PCA would sweep your problems!
What is reducing dimension? I will show you an example as follows: First, suppose you have a five-dimensional data set :
| Id | 1-d | 2-d | 3-d | 4-d | 5-d |
|---|---|---|---|---|---|
| data-1 | 1 | 2 | 3 | 4 | 5 |
| data-2 | 6 | 7 | 8 | 9 | 10 |
| .. | .. | .. | .. | .. |
Then, you could pick up PC1 and PC2 after PCA to reduce dimension for plotting:
| Id | PC1 | PC2 |
|---|---|---|
| data-1 | 0.3 | 0.6 |
| data-2 | 0.1 | 1.2 |
| .. | .. | .. |
PC1 and PC2 are the result obtained through data is projection on the unit vectors, which enable result to have the biggest variance(means its distribution is wide) and to be irrelevant(covariance = 0).
1.2 Algorithm#
- Normalize \(d\) dimension raw data
- Create the covariance matrix
- Calculate the eigenvalues of the covariance matrix and the corresponding eigenvectors
- The eigenvectors are sorted in the matrix according to the corresponding feature value, and the first k rows are formed into a matrix \(W\). (\(k<<d\))
- \(Y = xW\) is the result after reducing dimension to k dimension
Note: There are two prerequisites for conducting PCA
- Raw data has no NA
- The raw data should be normalized
2. PCA from scratch#
- Importing necessary modules
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
- Creating raw data
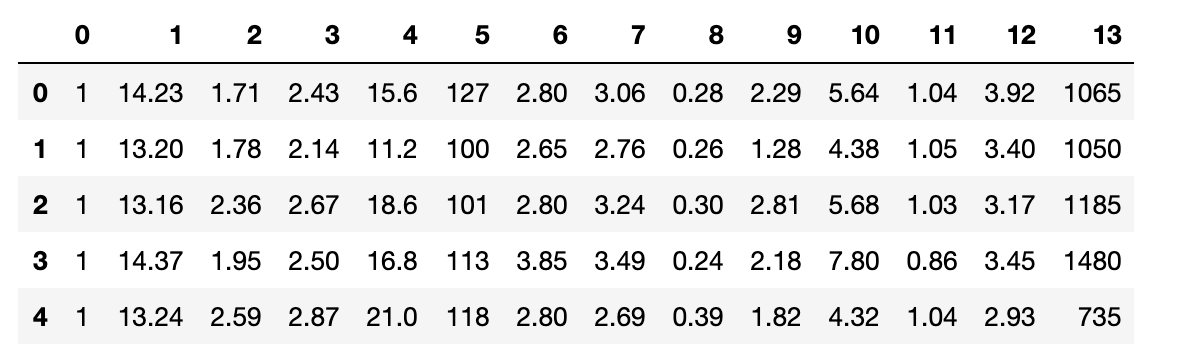
# get data set
df_wine = pd.read_csv(
"http://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data",
header=None,
engine="python",
)
# check data
df_wine.head()
- Creating train and test data set
# create train and test data set
X, y = df_wine.iloc[:, 1:], df_wine.iloc[:, 0]
x_train, x_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, stratify=y, random_state=0
)
- Standardizing the features
# create standard instance
sc = StandardScaler()
# standard data
x_train_std = sc.fit_transform(x_train)
x_test_std = sc.fit_transform(x_test)
- Creating the covariance matrix and Getting eigenvectors and eigenvalues
the calculation of the covariance matrix :
$$ \sigma_{jk} = \frac{1}{n} \sum^{n}_{i=1}\bigg(x_{j}^{(i)} - \mu_j\bigg)\bigg(x_{k}^{(i)} - \mu_k\bigg) $$
Then, using numpy.cov and numpy.linalg.eig to get the covariance matrix and eigenvectors respectively
# calculate the covariance matrix
cov_mat = np.cov(x_train_std.T)
# Getting eigenvectors and eigenvalues
eigen_vals, eigen_vecs = np.linalg.eig(cov_mat)
NOTE: there are 13 eigenvectors totally, the number of eigenvalues might be not as same as the number of features sometimes.
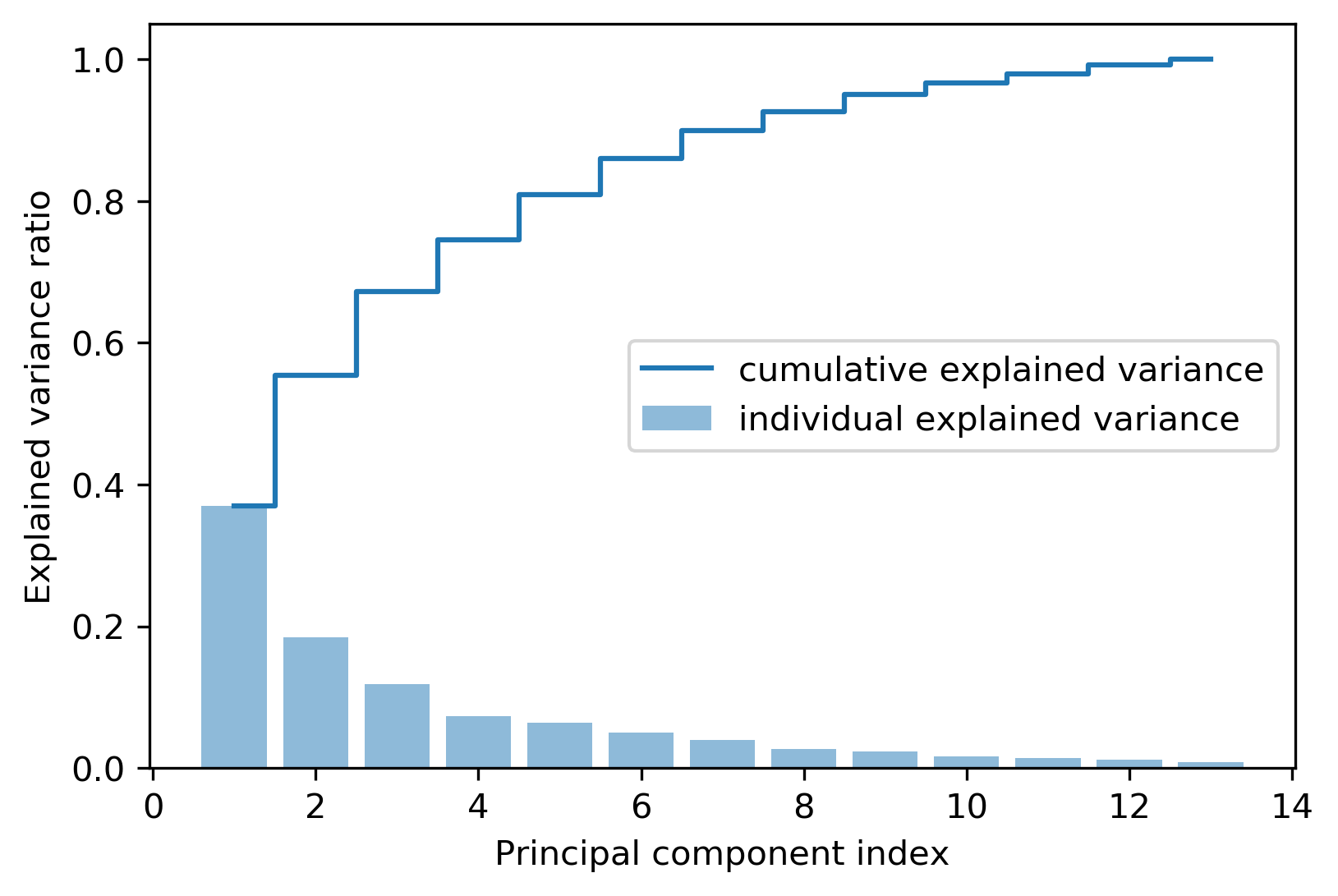
Firstly, plotting the Variance interpretation ratio, which is obtained through eigenvalue \(\lambda_j\) divided by the sum of all the eigenvalues:
$$ \frac{\lambda*j}{\sum^d_{j=1}\lambda_j} $$
# get sum of all the eigenvalues
tot = sum(eigen_vals)
# get variance interpretation ratio
var_exp = [(i / tot) for i in sorted(eigen_vals, reverse=True)]
cum_var_exp = np.cumsum(var_exp)
Besides, plotting the result to get in-depth understanding:
plt.figure() # create plot
# create bar plot
plt.bar(
range(1, 14),
var_exp,
alpha=0.5,
label="individual explained variance",
)
# create step plot
plt.step(range(1, 14), cum_var_exp, where="mid", label="cumulative explained variance")
# add label
plt.ylabel("Explained variance ratio")
plt.xlabel("Principal component index")
# add legend
plt.legend(loc="best")
# save picture
plt.savefig("pca_index.png", format="png", bbox_inches="tight", dpi=300)
- Selecting the first k values to form matrix \(W\)
# integrate eigenvalues and eigenvectors
eigen_paris = [
(np.abs(eigen_vals[i]), eigen_vecs[:, i]) for i in range(len(eigen_vals))
]
# sort according to eigenvalues
eigen_paris.sort(key=lambda x: x[0], reverse=True)
# pick up the first 2 eigenvalues
w = np.hstack([eigen_paris[0][1][:, np.newaxis], eigen_paris[1][1][:, np.newaxis]])
# check matrix x
w

- Transforming raw data
# reduce dimension
x_train_pca = x_train_std.dot(w)
# check resulted data
x_train_pca.shape
(124, 2)
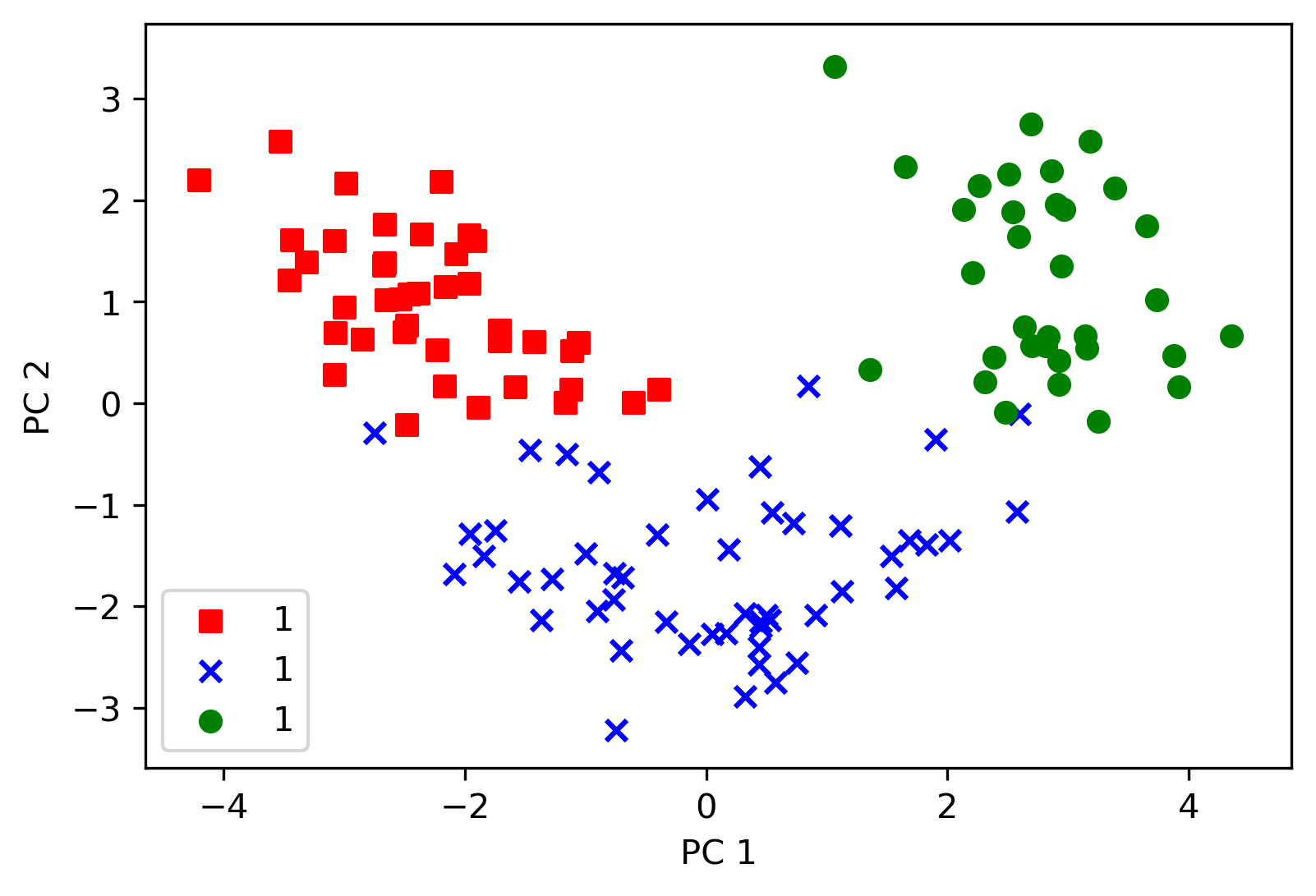
Then plotting the result and putting the label in terms of original info, but keeping in mind PCA is unsupervised learning skill without labels
# init colors and markers
colors = ["r", "b", "g"]
markers = ["s", "x", "o"]
# plot scatter
for l, c, m in zip(np.unique(y_train), colors, markers):
plt.scatter(
x_train_pca[y_train == l, 0],
x_train_pca[y_train == l, 1],
c=c,
label=1,
marker=m,
)
# add label and legend
plt.xlabel("PC 1")
plt.ylabel("PC 2")
plt.legend(loc="lower left")
plt.savefig("distribution.png", format="png", bbox_inches="tight", dpi=300)
3. PCA by scikit-learn#
we can conduct PCA easily by sklearn
- Importing modules
from sklearn.decomposition import PCA
from matplotlib.colors import ListedColormap
from sklearn.linear_model import LogisticRegression
- Defining function of plot_decision_region
def plot_dicision_regions(X, y, classifier, resolution=0.02):
# init markers and colors
markers = ("s", "x", "o", "^", "v")
colors = ("red", "blue", "lightgreen", "gray", "cyan")
cmap = ListedColormap(colors[: len(np.unique(y))])
# create info for plot region
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(
np.arange(x1_min, x1_max, resolution), np.arange(x2_min, x2_max, resolution)
)
# test classifier's accurate
z = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
z = z.reshape(xx1.shape)
# plot decision region
plt.contourf(xx1, xx2, z, alpha=0.4, cmap=cmap)
# set x,y length
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
# plot result
for idx, cl in enumerate(np.unique(y)):
plt.scatter(
x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.6,
color=cmap(idx),
edgecolor="black",
marker=markers[idx],
label=cl,
)
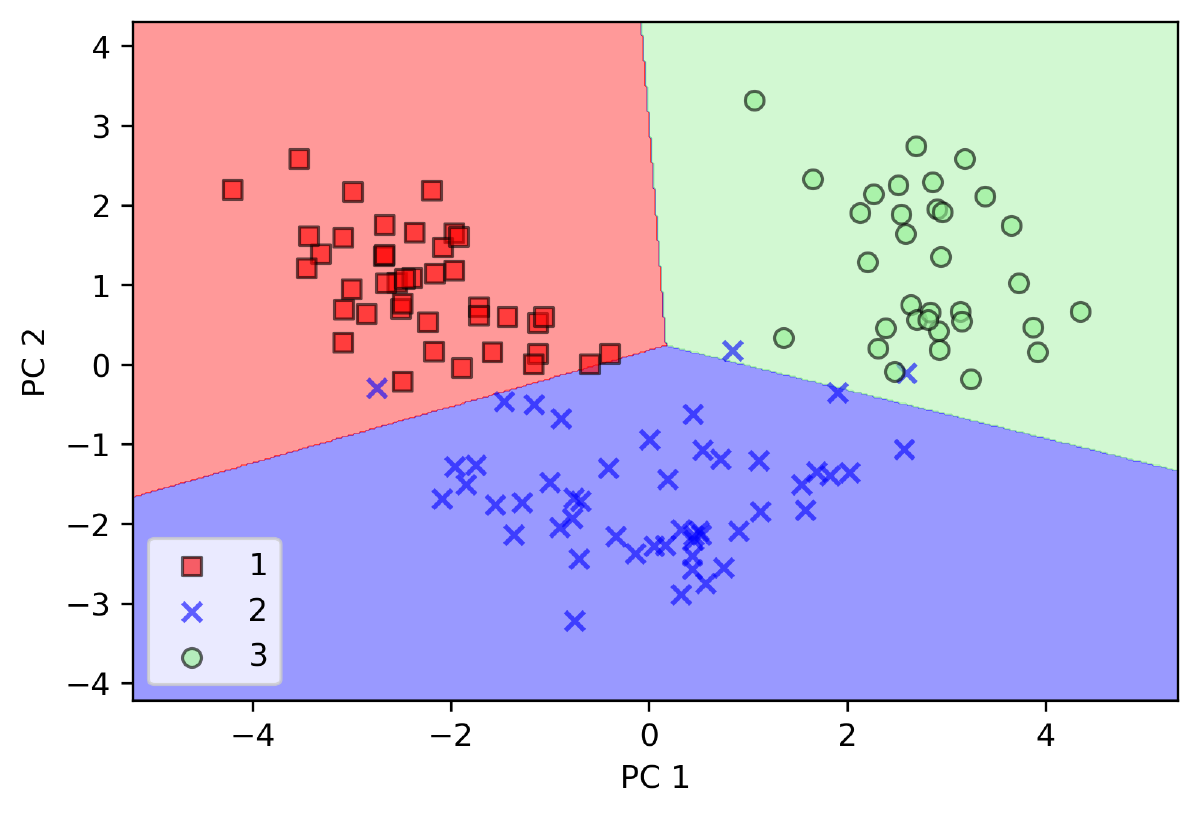
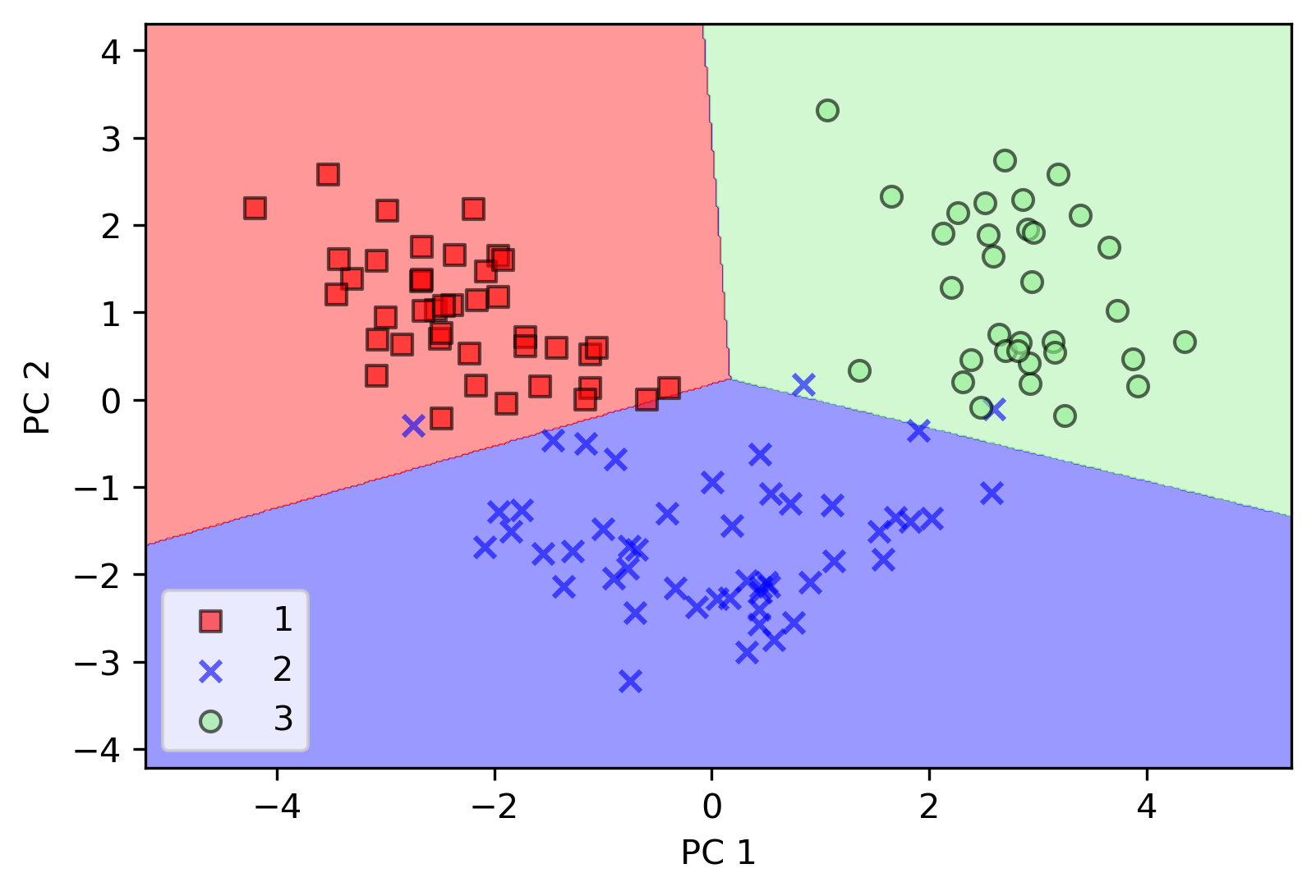
- PCA by sklearn
# create pca instance
pca = PCA(n_components=2)
# create classifier instance
lr = LogisticRegression()
# reduce dimension for data set
x_train_pca = pca.fit_transform(x_train_std)
x_test_pca = pca.transform(x_test_std)
# classify x_train_pca
lr.fit(x_train_pca, y_train)
# plot decision region
plot_dicision_regions(x_train_pca, y_train, classifier=lr)
# add info
plt.xlabel("PC 1")
plt.ylabel("PC 2")
plt.legend(loc="lower left")
plt.show()
TIPS:
You can set n_components = None, and the result would retain all principle components. Moreover, you could call explained_variance_ration_ to use variance explanation ratio.
3. Summary#
All the above are the main content, welcome everybody communicates with me! 🤠
Reference book : Python machine learning